Simple droughts workflow#
Click to launch this workflow on MyBinder.
Click 
This is the second and simpler drought workflow.
Risk assessment methodology#
The approach is to visualise the exposed vulnerable population to the drought. This can be done by overlying the hazard data of drought, expressed as Combined Drought Indicator (CDI), and population data.
Hazard data
The Combined Drought Indicator (CDI) that is implemented in the European Drought Observatory (EDO) is used to identify areas affected by agricultural drought, and areas with the potential to be affected. CDI can be downloaded from the Copernicus data server and is derived by combining three drought indicators produced operationally in the EDO framework - namely the
Standardized Precipitation Index (SPI),
the Soil Moisture Anomaly (SMA), and
the FAPAR Anomaly - in such a way that areas are classified according to three primary drought classes:
“Watch” - indicating that precipitation is less than normal;
“Warning” - indicating that soil moisture is in deficit; and
“Alert” - indicating that vegetation shows signs of stress.
Two additional classes - namely
“Partial recovery” and
“Recovery” - identify the stages of the vegetation recovery process.
Standardized Precipitation Index (SPI): The SPI indicator measures precipitation anomalies at a given location, based on a comparison of observed total precipitation amounts for an accumulation period of interest (e.g. 1, 3, 12, 48 months), with the long-term historic rainfall record for that period (McKee et al., 1993; Edwards and McKee, 1997).
Soil Moisture Anomaly (SMA): The SMA indicator is derived from anomalies of estimated daily soil moisture (or soil water) content - represented as standardized soil moisture index (SMI) - which is produced by the JRC’s LISFLOOD hydrological model, and which has been shown to be effective for drought detection purposes (Laguardia and Niemeyer, 2008).
FAPAR Anomaly: The FAPAR Anomaly indicator is computed as deviations of the biophysical variable Fraction of Absorbed Photosynthetically Active Radiation (FAPAR), composited for 10- day intervals, from long-term mean values. Satellite-measured FAPAR represents the fraction of incident solar radiation that is absorbed by land vegetation for photosynthesis, and is effective for detecting and assessing drought impacts on vegetation canopies (Gobron et al., 2005).
LEVEL |
COLOUR |
CLASIFICATION CONDITION |
|---|---|---|
Watch |
yellow |
SPI-3 <-1 or SPI-1 < -2 |
Warning |
orange |
SMA <-1 and (SPI-3 < -1 or SPI-1 < -2) |
Alert |
red |
\(\Delta\)FAPAR < -1 and (SPI-3 < -1 or SPI-1 < -2) |
Partial recovery |
brown |
(\(\Delta\)FAPAR < -1 and (SPI-3m-1 < -1 or SPI-3 > -1) or |
Full recovery |
grey |
(SPI-3m-1 < -1 and SPI-3 > -1) or |
Exposure data
Exposure is assessed by overlaying the European population density map at 30 arcsec resolution using the Global Human Settlement Population dataset dataset.
Preparation work#
Load libraries#
Find more info about the libraries used in this workflow here 👆
In this notebook we will use the following Python libraries:
os - To create directories and work with files
pooch - To download and unzip the data
rasterio - To access and explore geospatial raster data in GeoTIFF format
xarray - To process the data and prepare it for damage calculation
rioxarray - Rasterio xarray extension - to make it easier to use GeoTIFF data with xarray
cartopy - To plot the maps
matplotlib - For plotting as well
import os
import pooch
import numpy as np
import rasterio
from pathlib import Path
import rioxarray as rxr
import xarray as xr
import pyproj
import matplotlib as mpl
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
Create the directory structure#
In order for this workflow to work even if you download and use just this notebook, we need to set up the directory structure.
Next cell will create the directory called ‘drought_workflow’ in the same directory where this notebook is saved.
The cell after will create the data directory inside the drought_workflow.
workflow_folder = 'drought_workflow'
if not os.path.exists(workflow_folder):
os.makedirs(workflow_folder)
data_dir = os.path.join(workflow_folder,'data')
if not os.path.exists(data_dir):
os.makedirs(data_dir)
Download data#
In this workflow we will have a mix of data that is available to download from the notebook, and the data that has to be manually downloaded on the website.
Since there is no API to download this data, we can use pooch library to donwload and unzip it.
Pooch will check if the zip file already exists by comparing the hash of the file with what is stored in the default and only download it if it is not already there.
Population data#
First we need the information on the population. We will download it from the JRC data portal.
Once the data is downloaded and unzipped, Pooch will list the content of the directory with the data.
In this example we are using population data from JRC data portal - the Global Human Settlement Layer (GHSL). Note that this dataset is available in other resolutions and projections.
We are downloading global data, but it is also available in regional grid, by clicking on the grid box in the GHSL page.
url = 'https://jeodpp.jrc.ec.europa.eu/ftp/jrc-opendata/GHSL/GHS_POP_GLOBE_R2023A/GHS_POP_E2020_GLOBE_R2023A_4326_30ss/V1-0/GHS_POP_E2020_GLOBE_R2023A_4326_30ss_V1_0.zip'
pooch.retrieve(
url=url,
known_hash=None,
path=data_dir,
processor=pooch.Unzip(extract_dir=''))
The zip file contains data, as well as metadata and the documentation in the pdf file.
The Combined Drought Indicator (CDI)#
This dataset must be manually downloaded from the Copernicus Drought Observatory data server. Here we will use only data from 2022. However data is available for the period 2012-2023.
To download the data, you can select all years and click Download. Save the data in the data_dir folder.
Here we will use pooch to download the data for 2022.
url = 'https://jeodpp.jrc.ec.europa.eu/ftp/jrc-opendata/DROUGHTOBS/Drought_Observatories_datasets/EDO_Combined_Drought_Indicator/ver3-0-1/cdinx_m_euu_20220101_20221221_t.nc'
filename = 'cdinx_m_euu_20220101_20221221_t.nc'
pooch.retrieve(
url=url,
known_hash='7a40b16e5a0cfd53a22ef092f6a8bb511925972c9aaa9913a563c7e8585492e7',
path=data_dir,
fname=filename)
Note that now we have a directory drought_workflow/data where all the zip files and unzipped files are (or should be).
We can list all the files in the data_dir using the os library.
with os.scandir(data_dir) as entries:
for entry in entries:
print(entry.name)
Explore the data#
Now that we have downloaded and unpacked the needed data, we can have a look what is inside.
Population data#
Population data is in file with filename: GHS_POP_E2030_GLOBE_R2023A_54009_1000_V1_0.tif
We can use rioxaray to load this file.
filename_population = f'{data_dir}/GHS_POP_E2020_GLOBE_R2023A_4326_30ss_V1_0.tif'
population = rxr.open_rasterio(filename_population)
population
<xarray.DataArray (band: 1, y: 21384, x: 43202)>
[923831568 values with dtype=float64]
Coordinates:
* band (band) int64 1
* x (x) float64 -180.0 -180.0 -180.0 -180.0 ... 180.0 180.0 180.0
* y (y) float64 89.1 89.09 89.08 89.07 ... -89.08 -89.09 -89.1
spatial_ref int64 0
Attributes:
AREA_OR_POINT: Area
STATISTICS_MAXIMUM: 277608.74043128
STATISTICS_MEAN: 8.4874269494672
STATISTICS_MINIMUM: 0
STATISTICS_STDDEV: 235.7608994699
STATISTICS_VALID_PERCENT: 100
scale_factor: 1.0
add_offset: 0.0Explore the file content
Fell free to explore the content and structure of the dataset.
Note the coordinates, dimensions and attributes!
Find the information about spatial references, statistics👆 (click)
👋 Click on spatial_ref 📄 show/hide attributes to see the information about projection of this dataset.
👋 Click on Attributes to find the STATISTICS attributes including minimum, maximum and other statistics
One thing you might notice that is missing for this dataset is the units for the population data.
This information can be found in the technical documentation of the dataset.
We can read that values are expressed as decimals (Float) and represent the absolute number of inhabitants of the cell.
The Combined Drought Indicator (CDI)#
All the downloaded netcdf files are stored in our data_dir folder, with filenames starting with: cdinx_m_euu_, followed by the start and end date period, and ending with _t.nc.
First we can explore one of them.
cdi_nc = xr.open_dataset(f"{data_dir}/cdinx_m_euu_20220101_20221221_t.nc")
cdi_nc
<xarray.Dataset>
Dimensions: (time: 36, lat: 950, lon: 1000)
Coordinates:
* lat (lat) int64 5497500 5492500 5487500 ... 762500 757500 752500
* lon (lon) int64 2502500 2507500 2512500 ... 7487500 7492500 7497500
* time (time) datetime64[ns] 2022-01-01 2022-01-11 ... 2022-12-21
Data variables:
cdinx (time, lat, lon) float32 ...
3035 float32 ...
Attributes: (12/27)
Source_Software: dbinterface.py, dbexport.py, netcdf_handling.py
creator_name: Carolina Arias Munoz
Conventions: CF-1.6
_CoordSysBuilder: ucar.nc2.dataset.conv.CF1Convention
date_created: 2023-06-23
01.title: Combined Drought Indicator (CDI), v.3.0.1
... ...
17.factsheet_url: https://edo.jrc.ec.europa.eu/documents/factsh...
18.jrc_data_catalogue_url: https://data.jrc.ec.europa.eu/dataset/afa8a5e...
19.sample_url: /images/map_examples/dbio_data_previews/cdinx...
20.metadata_last_updated: 2023-04-05
21.values_legend: 0: No drought, 1: Watch. Precipitation defici...
22.version_notes: Current version: Version 3.0.1 covers data fr...We can notice that there are 36 time stamps for 12 months in the year 2022.
This is because this index is caluclated from the satellite data that takes 10 days to collect the information over the globe. Each time step has index calculated over the previous 10 days.
Tip
Explore the file content
Fell free to explore the content and structure of the datasets.
Note the coordinates, dimensions and attributes!
Hint
Find the information about spatial references, statistics👆 (click)
👋 Click on 3035 📄 show/hide attributes to see the information about projection of this dataset.
👋 Click on Attributes to find the extensive metadata including projection, resolution, extent, links to documentation, data catalogue etc
Process the data#
In this workflow we want to overlay the population and drought data, to have a better understanding where are densely populated area affected by the drought.
Hint
Take a closer look at the dimensions and coordinates of our two data objects.
Notice that population data is xarray data array and CDI is a dataset. The difference is that dataset contains two variables, the cdinx which is the data variable, and 3035 which has the informaiton about the projection.
Notice that population data has x and y as spatial dimensions, while CDI has lat and lon
The projections and resolutions are very different as well, but they are both in meters.
The CDI data has time dimension
To be able to plot these two datasets together we must have them in the same projection and zoom to the same area.
For now we select just one time stamp for the CDI data, for start. We can use xarray sel() method to select one time.
To check which times are available you can ispect the data few cells above, or print it like in the next cell.
cdi_nc.time
<xarray.DataArray 'time' (time: 36)>
array(['2022-01-01T00:00:00.000000000', '2022-01-11T00:00:00.000000000',
'2022-01-21T00:00:00.000000000', '2022-02-01T00:00:00.000000000',
'2022-02-11T00:00:00.000000000', '2022-02-21T00:00:00.000000000',
'2022-03-01T00:00:00.000000000', '2022-03-11T00:00:00.000000000',
'2022-03-21T00:00:00.000000000', '2022-04-01T00:00:00.000000000',
'2022-04-11T00:00:00.000000000', '2022-04-21T00:00:00.000000000',
'2022-05-01T00:00:00.000000000', '2022-05-11T00:00:00.000000000',
'2022-05-21T00:00:00.000000000', '2022-06-01T00:00:00.000000000',
'2022-06-11T00:00:00.000000000', '2022-06-21T00:00:00.000000000',
'2022-07-01T00:00:00.000000000', '2022-07-11T00:00:00.000000000',
'2022-07-21T00:00:00.000000000', '2022-08-01T00:00:00.000000000',
'2022-08-11T00:00:00.000000000', '2022-08-21T00:00:00.000000000',
'2022-09-01T00:00:00.000000000', '2022-09-11T00:00:00.000000000',
'2022-09-21T00:00:00.000000000', '2022-10-01T00:00:00.000000000',
'2022-10-11T00:00:00.000000000', '2022-10-21T00:00:00.000000000',
'2022-11-01T00:00:00.000000000', '2022-11-11T00:00:00.000000000',
'2022-11-21T00:00:00.000000000', '2022-12-01T00:00:00.000000000',
'2022-12-11T00:00:00.000000000', '2022-12-21T00:00:00.000000000'],
dtype='datetime64[ns]')
Coordinates:
* time (time) datetime64[ns] 2022-01-01 2022-01-11 ... 2022-12-21
Attributes:
standard_name: timecdi = cdi_nc.sel(time='2022-08-01T00:00:00.000000000')
Select the area of interest#
In this workflow we will concentrate on the area of Catalonia. It roughly between 0°E and 3.4°E longitude and 40.5°N and 42.9°N latidude.
However, the CDI dataset has the spatial coordinates in meters, so it is not easy to know the values to which we need to clip the area.
For this we can use the pyproj library.
Here we use the proj-string of the Lambert Azimuthal Equal Area projection. This information can be found in the attributes of the CDI datatset under ‘3035’ variable.
cdi_nc["3035"].proj4_params
'+proj=laea +lat_0=52 +lon_0=10 +x_0=4321000+y_0=3210000 +ellps=GRS80 +units=m +no_defs'
xmin=0
ymin=40.5
xmax=3.4
ymax=42.9
xyproj = pyproj.Proj('+proj=laea +lat_0=52 +lon_0=10 +x_0=4321000 +y_0=3210000 +ellps=GRS80 +units=m +no_defs')
latlonproj = pyproj.Proj('epsg:4326')
transformer = pyproj.Transformer.from_proj(latlonproj, xyproj)
(xmin_xyproj,ymin_xyproj) = transformer.transform(ymin, xmin)
(xmax_xyproj,ymax_xyproj) = transformer.transform(ymax, xmax)
Here are our transformed coordinates of the boundary box.
xmin_xyproj,ymin_xyproj
(3472008.40051275, 1989781.4970224823)
xmax_xyproj,ymax_xyproj
(3781134.3733767485, 2222944.719518429)
Caution
Attention!
One very sneaky thing to note here is that latitudes (y dimension) in both datasets is decreasing.
This is very important as this means we need to have the minimum and maximum y value in this order when selecting the area.
There are two ways of selecting area. We can use xarray sel() function or rasterio rio.clip_box.
catalonia_pop = population.rio.clip_box(
minx=xmin,
miny=ymin,
maxx=xmax,
maxy=ymax,
crs="EPSG:3050",
)
catalonia_population = population.sel(y=slice(ymax,ymin), x=slice(xmin,xmax))
catalonia_population = population.sel(y=slice(ymax,ymin),
x=slice(xmin,xmax))
catalonia_population
<xarray.DataArray (band: 1, y: 288, x: 408)>
[117504 values with dtype=float64]
Coordinates:
* band (band) int64 1
* x (x) float64 0.004583 0.01292 0.02125 ... 3.38 3.388 3.396
* y (y) float64 42.9 42.89 42.88 42.87 ... 40.53 40.52 40.51 40.5
spatial_ref int64 0
Attributes:
AREA_OR_POINT: Area
STATISTICS_MAXIMUM: 277608.74043128
STATISTICS_MEAN: 8.4874269494672
STATISTICS_MINIMUM: 0
STATISTICS_STDDEV: 235.7608994699
STATISTICS_VALID_PERCENT: 100
scale_factor: 1.0
add_offset: 0.0catalonia_cdi = cdi.sel(lat=slice(ymax_xyproj,ymin_xyproj),
lon=slice(xmin_xyproj,xmax_xyproj))
catalonia_cdi
<xarray.Dataset>
Dimensions: (lat: 47, lon: 62)
Coordinates:
* lat (lat) int64 2222500 2217500 2212500 ... 2002500 1997500 1992500
* lon (lon) int64 3472500 3477500 3482500 ... 3767500 3772500 3777500
time datetime64[ns] 2022-08-01
Data variables:
cdinx (lat, lon) float32 ...
3035 float32 ...
Attributes: (12/27)
Source_Software: dbinterface.py, dbexport.py, netcdf_handling.py
creator_name: Carolina Arias Munoz
Conventions: CF-1.6
_CoordSysBuilder: ucar.nc2.dataset.conv.CF1Convention
date_created: 2023-06-23
01.title: Combined Drought Indicator (CDI), v.3.0.1
... ...
17.factsheet_url: https://edo.jrc.ec.europa.eu/documents/factsh...
18.jrc_data_catalogue_url: https://data.jrc.ec.europa.eu/dataset/afa8a5e...
19.sample_url: /images/map_examples/dbio_data_previews/cdinx...
20.metadata_last_updated: 2023-04-05
21.values_legend: 0: No drought, 1: Watch. Precipitation defici...
22.version_notes: Current version: Version 3.0.1 covers data fr...The last thing left to do is reproject the CDI data.
In order for rioxarray to be able to reproject the data, it needs to know the projection. NetCDF files store the projection information in its variables or attributes, so for rioxarray we need to write it using rio.write_crs().
More info about coordinate system and projections in NetCDF data👆 (click)
Read more about projections and coordinate information in NetCDF data in:
CF convention and in
catalonia_cdi.rio.write_crs(3035, inplace=True)
catalonia_cdi_latlon = catalonia_cdi.rio.reproject("EPSG:4326")
Plot it together#
Basic plot#
With a little bit of code, we can simply plot these two datasets together
catalonia_cdi_latlon.cdinx.plot(cmap=plt.cm.PuRd, extend='max')
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000, 45000]
catalonia_population.plot(cmap=plt.cm.BuGn, levels=p_levels, extend='max')
<matplotlib.collections.QuadMesh at 0x1923b1cd0>
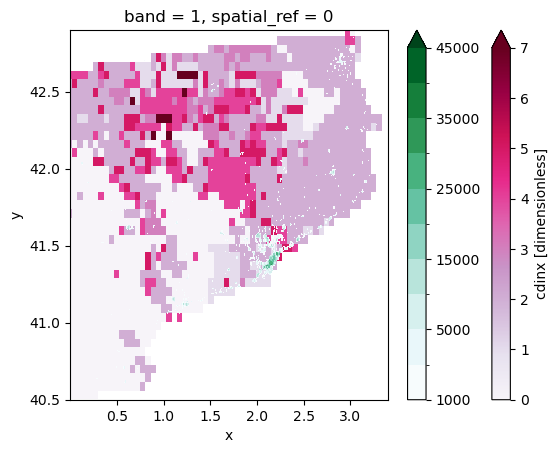
This plot doesn’t really help us doing climate risk assessemnt, so we can try to make a few changes to the map.
Custom plot#
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
ax.stock_img()
# Plot CDI data
cmap = mpl.colors.ListedColormap(["#ffffff", "#f0e442", "#f5aa00", "#dc050c","#0072b2","#cc79a7","#009e73", "#c8c8c8"])
c = catalonia_cdi_latlon.cdinx.plot(ax=ax, cmap=cmap, vmin=-0.5, vmax=7.5, add_colorbar=False, add_labels=False)
#Plot the population
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000]
p = catalonia_population.plot(ax=ax, cmap=plt.cm.BuGn, levels=p_levels, extend='max', add_colorbar=False, add_labels=False)
# Customize gridlines
gridlines = p.axes.gridlines(draw_labels=True, dms=False, xlocs=[0, 2, 4], ylocs=[40, 42])
ax.add_feature(cfeature.BORDERS)
ax.add_feature(cfeature.COASTLINE)
ax.add_feature(cfeature.RIVERS)
plt.title('The Combined Drought Indicator and Population number', loc = "left")
#population colorbar
cax_p = fig.add_axes([ax.get_position().x0,
ax.get_position().y0-0.03,
ax.get_position().width,
0.02])
cbar_p = plt.colorbar(p, orientation="horizontal", cax=cax_p, ticks=[1000, 5000, 15000, 25000, 35000])
cbar_p.ax.tick_params(labelsize=8)
# CDI colorbar
cax_c = fig.add_axes([ax.get_position().x0,
ax.get_position().y0,
ax.get_position().width,
0.08])
cbar_c = plt.colorbar(c, orientation="horizontal", cax=cax_c)
cbar_c.ax.get_xaxis().set_ticks([])
for j, lab in enumerate(['No drought', 'Watch:\n rainfall\n deficit', 'Warning:\n soil moisture\n deficit', 'Alert','Recovery','Temporary\n soil moisture\n recovery','Temporary\n fAPAR\n recovery','No data']):
cbar_c.ax.text(j, .5, lab, ha='center', va='center', size=8.5)
#text
barcelona_lon, barcelona_lat = 2.1686, 41.3874
text_kwargs = dict(fontsize=12, color='#000032', transform=ccrs.PlateCarree())
p.axes.text(barcelona_lon, barcelona_lat, '+ Barcelona', horizontalalignment='left', **text_kwargs)
Text(2.1686, 41.3874, '+ Barcelona')
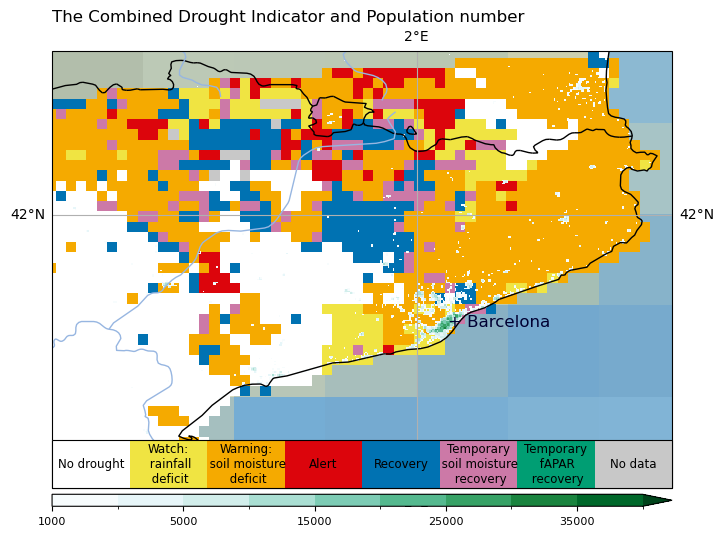
Conclusions#
In this workflow, we have seen how to explore, process and visualise the data needed to analyse drought and population data.
Contributors#
Milana Vuckovic, ECMWF
Maurizio Mazzoleni, Vrije Universiteit Amsterdam
Appendix I - Customised plot explained step by step#
We will:
Add coastlines, rivers and country borders using cartopy
Set distinct colour levels for CDI
Remove the default colorbar and labels
Add custom colorbars and title
Add custom gridlines
And finally add a label for Barcelona
Background#
First we make a basic plot and set geographical extent to our domain. This way we can zoom in without having to manipulate the data.
We can also add some (low resolution) background.
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
# Add background image with orography
ax.stock_img()
<matplotlib.image.AxesImage at 0x19267ec50>
Adding the data#
Next we add the data.
Here we customize the levels on both plots. We will also remove the default colorbar and labels, so in the next step we can add our own.
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
ax.stock_img()
#----------- New code to plot the data -----------#
# Plot CDI data
cmap = mpl.colors.ListedColormap(["#ffffff", "#f0e442", "#f5aa00", "#dc050c","#0072b2","#cc79a7","#009e73", "#c8c8c8"])
c = catalonia_cdi_latlon.cdinx.plot(ax=ax, cmap=cmap, vmin=-0.5, vmax=7.5, add_colorbar=False, add_labels=False)
#Plot the population
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000]
p = catalonia_population.plot(ax=ax, cmap=plt.cm.BuGn, levels=p_levels, extend='max', add_colorbar=False, add_labels=False)
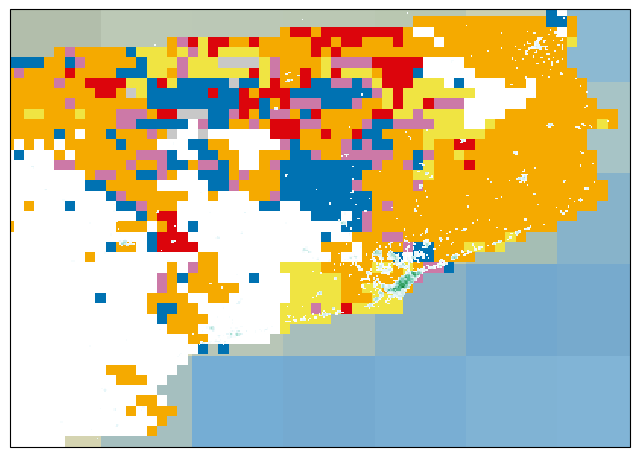
Adding the geographical features#
Next we add the gridlines, coastlines, rivers and country borders.
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
ax.stock_img()
# Plot CDI data
cmap = mpl.colors.ListedColormap(["#ffffff", "#f0e442", "#f5aa00", "#dc050c","#0072b2","#cc79a7","#009e73", "#c8c8c8"])
c = catalonia_cdi_latlon.cdinx.plot(ax=ax, cmap=cmap, vmin=-0.5, vmax=7.5, add_colorbar=False, add_labels=False)
#Plot the population
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000]
p = catalonia_population.plot(ax=ax, cmap=plt.cm.BuGn, levels=p_levels, extend='max', add_colorbar=False, add_labels=False)
#----------- New code to customize gridlines -----------#
gridlines = p.axes.gridlines(draw_labels=True, dms=False, xlocs=[0, 2, 4], ylocs=[40, 42])
ax.add_feature(cfeature.BORDERS)
ax.add_feature(cfeature.COASTLINE)
ax.add_feature(cfeature.RIVERS)
<cartopy.mpl.feature_artist.FeatureArtist at 0x19251c3d0>
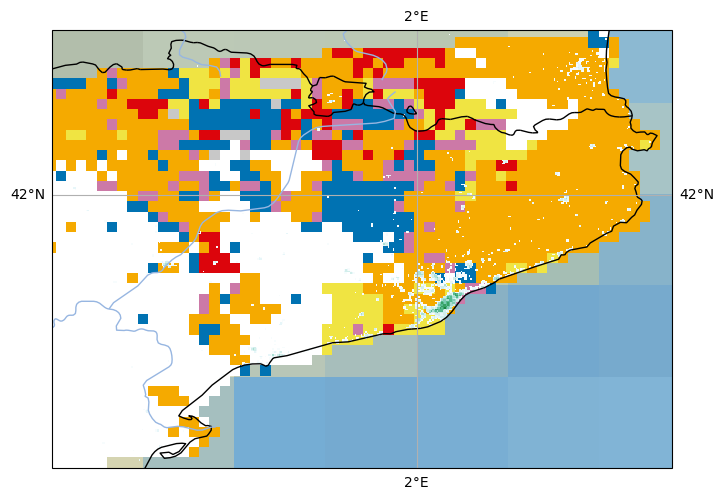
Adding the title#
Then we add the title.
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
ax.stock_img()
# Plot CDI data
cmap = mpl.colors.ListedColormap(["#ffffff", "#f0e442", "#f5aa00", "#dc050c","#0072b2","#cc79a7","#009e73", "#c8c8c8"])
c = catalonia_cdi_latlon.cdinx.plot(ax=ax, cmap=cmap, vmin=-0.5, vmax=7.5, add_colorbar=False, add_labels=False)
#Plot the population
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000]
p = catalonia_population.plot(ax=ax, cmap=plt.cm.BuGn, levels=p_levels, extend='max', add_colorbar=False, add_labels=False)
# Customize gridlines
gridlines = p.axes.gridlines(draw_labels=True, dms=False, xlocs=[0, 2, 4], ylocs=[40, 42])
ax.add_feature(cfeature.BORDERS)
ax.add_feature(cfeature.COASTLINE)
ax.add_feature(cfeature.RIVERS)
#----------- New code to add the title -----------#
plt.title('The Combined Drought Indicator and Population number', loc = "left")
Text(0.0, 1.0, 'The Combined Drought Indicator and Population number')
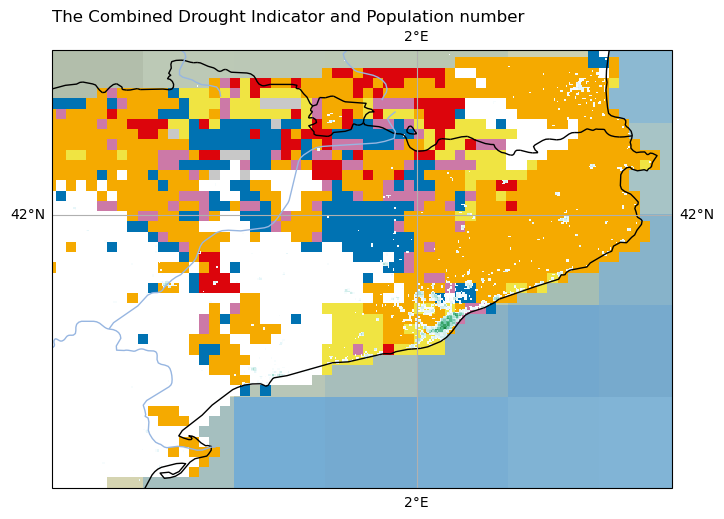
Adding the colorbar#
It is a little bit tricky to add colorbars to where we want them.
cax_p and cax_c variable define the positions and size of the colorbars. They get the default position and size of the colorbars and make them half as long and next to each other.
In the end we also make the tick labels smaller.
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
ax.stock_img()
# Plot CDI data
cmap = mpl.colors.ListedColormap(["#ffffff", "#f0e442", "#f5aa00", "#dc050c","#0072b2","#cc79a7","#009e73", "#c8c8c8"])
c = catalonia_cdi_latlon.cdinx.plot(ax=ax, cmap=cmap, vmin=-0.5, vmax=7.5, add_colorbar=False, add_labels=False)
#Plot the population
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000]
p = catalonia_population.plot(ax=ax, cmap=plt.cm.BuGn, levels=p_levels, extend='max', add_colorbar=False, add_labels=False)
# Customize gridlines
gridlines = p.axes.gridlines(draw_labels=True, dms=False, xlocs=[0, 2, 4], ylocs=[40, 42])
ax.add_feature(cfeature.BORDERS)
ax.add_feature(cfeature.COASTLINE)
ax.add_feature(cfeature.RIVERS)
plt.title('The Combined Drought Indicator and Population number', loc = "left")
#----------- New code to add the horizontal colorbars -----------#
cax_p = fig.add_axes([ax.get_position().x0,
ax.get_position().y0-0.03,
ax.get_position().width,
0.02])
cbar_p = plt.colorbar(p, orientation="horizontal", cax=cax_p, ticks=[1000, 5000, 15000, 25000, 35000])
cbar_p.ax.tick_params(labelsize=8)
# CDI colorbar
cax_c = fig.add_axes([ax.get_position().x0,
ax.get_position().y0,
ax.get_position().width,
0.08])
cbar_c = plt.colorbar(c, orientation="horizontal", cax=cax_c)
cbar_c.ax.get_xaxis().set_ticks([])
for j, lab in enumerate(['No drought', 'Watch:\n rainfall\n deficit', 'Warning:\n soil moisture\n deficit', 'Alert','Recovery','Temporary\n soil moisture\n recovery','Temporary\n fAPAR\n recovery','No data']):
cbar_c.ax.text(j, .5, lab, ha='center', va='center', size=8.5)
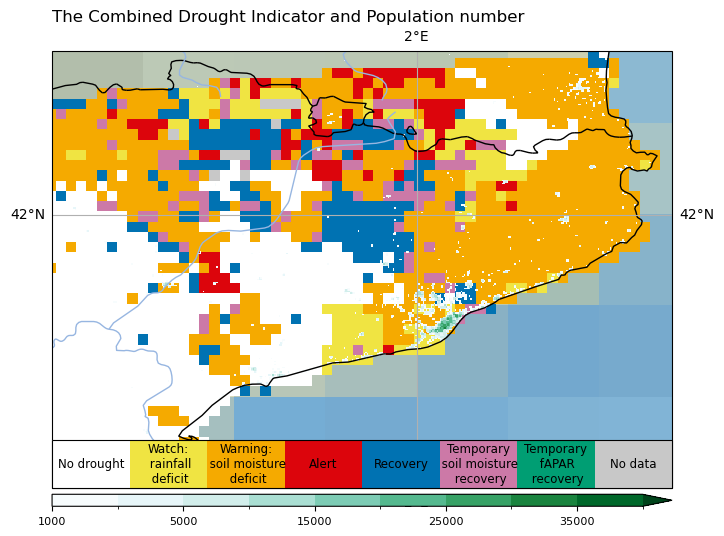
Adding the custom text label#
Finally we can add the label for Barcelona.
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(1, 1, 1, projection=ccrs.PlateCarree())
ax.set_extent([xmin, xmax, ymin, ymax], crs=ccrs.PlateCarree())
ax.stock_img()
# Plot CDI data
cmap = mpl.colors.ListedColormap(["#ffffff", "#f0e442", "#f5aa00", "#dc050c","#0072b2","#cc79a7","#009e73", "#c8c8c8"])
c = catalonia_cdi_latlon.cdinx.plot(ax=ax, cmap=cmap, vmin=-0.5, vmax=7.5, add_colorbar=False, add_labels=False)
#Plot the population
p_levels = [ 1000, 2500, 5000, 10000, 15000, 20000, 25000, 30000, 35000, 40000]
p = catalonia_population.plot(ax=ax, cmap=plt.cm.BuGn, levels=p_levels, extend='max', add_colorbar=False, add_labels=False)
# Customize gridlines
gridlines = p.axes.gridlines(draw_labels=True, dms=False, xlocs=[0, 2, 4], ylocs=[40, 42])
ax.add_feature(cfeature.BORDERS)
ax.add_feature(cfeature.COASTLINE)
ax.add_feature(cfeature.RIVERS)
plt.title('The Combined Drought Indicator and Population number', loc = "left")
cax_p = fig.add_axes([ax.get_position().x0,
ax.get_position().y0-0.03,
ax.get_position().width,
0.02])
cbar_p = plt.colorbar(p, orientation="horizontal", cax=cax_p, ticks=[1000, 5000, 15000, 25000, 35000])
cbar_p.ax.tick_params(labelsize=8)
# CDI colorbar
cax_c = fig.add_axes([ax.get_position().x0,
ax.get_position().y0,
ax.get_position().width,
0.08])
cbar_c = plt.colorbar(c, orientation="horizontal", cax=cax_c)
cbar_c.ax.get_xaxis().set_ticks([])
for j, lab in enumerate(['No drought', 'Watch:\n rainfall\n deficit', 'Warning:\n soil moisture\n deficit', 'Alert','Recovery','Temporary\n soil moisture\n recovery','Temporary\n fAPAR\n recovery','No data']):
cbar_c.ax.text(j, .5, lab, ha='center', va='center', size=8.5)
#----------- New code to add the text annotation -----------#
barcelona_lon, barcelona_lat = 2.1686, 41.3874
text_kwargs = dict(fontsize=12, color='#000032', transform=ccrs.PlateCarree())
p.axes.text(barcelona_lon, barcelona_lat, '+ Barcelona', horizontalalignment='left', **text_kwargs)
Text(2.1686, 41.3874, '+ Barcelona')

